Scientific Review
Innovative Antiviral Strategy Targeting PLpro: Discovery of Jun12682 and Analysis of Its Antipandemic Effects 
 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Medical Science, 2024, Vol. 14, No. 1 doi: 10.5376/ijmms.2024.14.0008
Received: 29 Mar., 2024 Accepted: 03 Apr., 2024 Published: 09 Apr., 2024
Xuan J., 2024, Innovative antiviral strategy targeting PLpro: Discovery of Jun12682 and analysis of its antipandemic effects, International Journal of Molecular Medical Science, 14(1): 56-60 (doi: 10.5376/ijmms.2024.14.0008)
The paper "Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model," authored by Bin Tan, Xiaoming Zhang, Ahmadullah Ansari, Prakash Jadhav, et al., was published in Science on March 29, 2024. The authors are affiliated with Department of Medicinal Chemistry, Ernest Mario School of Pharmacy, Rutgers, the State University of New Jersey, and Department of Physiological Sciences, College of Veterinary Medicine, Oklahoma State University, among others. This research focuses on a key enzyme of SARS-CoV-2, the papain-like protease (PLpro), which plays a crucial role in the virus's replication and immune evasion mechanisms. Using a structure-guided approach, the research team designed a series of non-covalent PLpro inhibitors. These inhibitors specifically target a newly discovered ubiquitin-binding site, Val70Ub, on PLpro, demonstrating their antiviral activity in both in vitro and in vivo models. The lead compound, Jun12682, showed excellent antiviral effects in a mouse model, improving survival rates, and reducing pulmonary viral loads, offering a new strategic direction for the treatment of SARS-CoV-2.
1 Experimental Data Analysis
In the study, the discovery and evaluation of Jun12682 were accomplished through a series of meticulous experimental steps. In vitro experiments demonstrated that Jun12682 has very strong inhibitory activity against PLpro, with an inhibitory constant (Ki) of only 37.7 nanomolar, and an effective concentration for 50% of maximal effect (EC50) value of 1.1 micromolar in the FlipGFP PLpro cell assay, indicating its high efficacy in inhibiting the key enzyme activity of the virus. Further in vivo experiments conducted in a mouse model showed that Jun12682 significantly improved the survival rate of mice infected with SARS-CoV-2, mitigated the virus-induced weight loss, and effectively reduced the viral load in the lungs. These experimental data collectively validate the value of Jun12682 as a potential anti-SARS-CoV-2 therapeutic drug.
Figure 1 displays the X-ray crystal structure of the hybrid covalent inhibitor Jun11313, designed based on XR8-24 and Cp7, with SARS-CoV-2 PLpro. In structure (B), the interactions between Jun11313 and the PLpro binding site are represented by hydrogen bonds (black dashed lines), with the spatial structure of Jun11313 shown in green ball-and-stick model. Figure (C) overlays the structure of PLpro with Jun11313 against the XR8-24 complex, highlighting the amino acid residues related to the binding of both compounds. Structure (D) further presents an overlay of the binding modes of Jun11313, XR8-24, and ubiquitin on PLpro. Structure (E) provides the generic chemical structure of the designed biarylphenyl PLpro inhibitors and highlights key interactions. Lastly, the flowchart (F) shows the optimization process for PLpro inhibitors, with Jun12682 being selected as the leading candidate for in vivo studies.
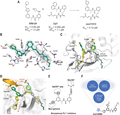 Figure 1 X-ray crystal structure of the covalent inhibitor Jun11313 with SARS-CoV-2 PLpro and structure-based design of biarylphenyl SARS-CoV-2PLpro inhibitors |
Figure 2 showcases a series of representative compounds of biarylbenzamide class SARS-CoV-2 PLpro inhibitors. Item A in the illustration represents the positive control GRL0617; items B, C to F are PLpro inhibitors containing thiophene and pyrazole. These compounds were characterized for their half-maximal inhibitory concentration (IC50) and inhibitory constant (Ki) through FRET-based enzymatic activity assays, and their cytotoxicity (CC50) in Vero E6 cells was evaluated. Moreover, the EC50 values for FlipGFP PLpro were tested using 293T cells transfected with PLpro and FlipGFP. The antiviral activity (EC50) of these compounds against SARS-CoV-2 was assessed in Caco-2 cell lines expressing hACE2 and hTMPRSS2. The figure also provides data on the half-life (T1/2) of PLpro inhibitors in mouse liver microsomes, with all data presented as the mean ± standard deviation of three technical replicates.
 Figure 2 Representative biarylbenzamide series of SARS-CoV-2 PLpro inhibitors |
Figure 3 displays the antiviral activity of PLpro inhibitors against different variants of SARS-CoV-2 and the mechanistic studies of Jun12682. Figures A, B, and C show the inhibitory effects of nirmatrelvir, Jun11941, and Jun12682 on the original WA1 strain, the Delta variant, the Omicron variant, and resistant strains, respectively. Jun12682 exhibits significant antiviral activity against all viral strains, with particularly strong effects against the Omicron variant. Figures D to G present the quantitative results of Jun12682 inhibiting SARS-CoV-2 PLpro enzymatic activity, including the inhibition of ubiquitin (Ub-AMC) and interferon-stimulated gene 15 (ISG15-AMC) hydrolysis, demonstrating its potent inhibitory action. Figures H and I are screening for inhibition against human proteases USP7 and USP14, where Jun12682 shows selective inhibition of PLpro over USP7 and USP14. Figure J evaluates the impact of Jun12682 and the control GRL0617 on the stability of SARS-CoV-2 PLpro through differential scanning fluorimetry, indicating that Jun12682 can effectively stabilize the PLpro protein. Overall, these data suggest that Jun12682 is a potent SARS-CoV-2 PLpro inhibitor with potential as a new antiviral drug.
 Figure 3 Antiviral activity of PLpro inhibitors against SARS-CoV-2 variants and mechanistic studies of Jun12682 |
Figure 4 illustrates the X-ray crystal structures of SARS-CoV-2 PLpro with multiple biarylbenzamide inhibitors, with a particular emphasis on the atomic model of Jun12682 at the PLpro binding site. This model reveals the hydrogen bonds (black dashed lines), van der Waals contacts (red dashed lines), and π-π interactions (light green dashed lines) between Jun12682 and PLpro. Additionally, for each compound, the inhibitory constant Ki from FRET enzymatic assays and the antiviral activity EC50 value against SARS-CoV-2 in Caco-2 cells are provided. The "polder map" of the inhibitors, displayed as a gray mesh, reveals the precise morphology of binding to PLpro, confirming the potential efficacy of these molecules in blocking viral functions.
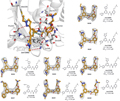 Figure 4 X-ray crystal structures of SARS-CoV-2 PLpro with biarylphenyl PLpro inhibitors |
Figure 5 presents the pharmacokinetic (PK) characteristics of PLpro inhibitors in vitro and in vivo and the in vivo antiviral efficacy of Jun12682. Figures A and B show the plasma concentration of different compounds in mice. Notably, Jun12682 exhibits favorable pharmacokinetic properties after oral administration (Figure C). Figure D summarizes the in vitro pharmacokinetic parameters of Jun12682, such as microsomal stability and CYP inhibition. Figures E to G and H to J designed treatment experiments for 5 days and 3 days, respectively, to evaluate the efficacy of Jun12682 against the SARS-CoV-2 N501Y variant infection in a mouse model, including changes in body weight and survival rates. Mice treated with Jun12682 showed lower viral loads (Figure K), reduced pathological damage (Figures N and O), and decreased levels of inflammatory factors (Figure M) compared to untreated mice. These results collectively suggest that Jun12682 has potential as an oral antiviral drug against SARS-CoV-2.
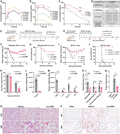 Figure 5 In vitro and in vivo PK profiling of PLpro inhibitors and in vivo antiviral efficacy of Jun12682 |
2 Analysis of Research Findings
In this study, the research team made a breakthrough confirmation that the papain-like protease (PLpro) of SARS-CoV-2 is an effective drug target. Through detailed structural analysis, they discovered the Val70Ub site on PLpro for the first time, suggesting its potential as a drug-binding site. This discovery provides a foundation for designing novel antiviral drugs. Using a structure-guided approach, the team successfully developed a series of non-covalent PLpro inhibitors targeting this new site. Among these inhibitors, Jun12682 was particularly noted for its exceptional efficacy. It not only demonstrated potent PLpro inhibitory activity in vitro but also proved to effectively inhibit SARS-CoV-2 replication in a mouse model, showing its great potential as a therapeutic drug. The discovery of Jun12682 validates the strategy of effectively blocking SARS-CoV-2 replication by targeting the Val70Ub site on PLpro, and it opens new directions for future drug development against SARS-CoV-2 and potential variants of coronaviruses. This breakthrough holds significant importance for current COVID-19 treatment research and offers new insights and strategies for the global scientific community in exploring more effective methods for viral treatment.
3 Evaluation of the Research
This study, by integrating advanced methods from structural biology and medicinal chemistry, successfully identified Jun12682 as a potential drug candidate against the SARS-CoV-2 virus, demonstrating the research team's exceptional capability in scientific innovation. Through precise analysis of the structure and function of the PLpro enzyme, the research not only deepened our understanding of viral biology but also showed how to translate complex scientific knowledge into practical therapeutic strategies. The discovery of Jun12682 validates the feasibility, both theoretically and practically, of therapies targeting the SARS-CoV-2 PLpro enzyme, offering new hope and possibilities for combating the current pandemic and potential future coronavirus variants.
4 Conclusions
Jun12682, as a novel PLpro inhibitor, has shown significant anti-SARS-CoV-2 activity both in vitro and in vivo, offering not only a new potential means to combat the COVID-19 pandemic but also new directions and strategies for the development of therapeutic drugs for coronavirus diseases. This work highlights the importance of PLpro as a drug target and demonstrates the power of structure-guided design methods in modern drug discovery.
5 Access the Full Text
Bin Tan et al., Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science 383, 1434-1440 (2024). DOI:10.1126/science.adm9724
Acknowledgments
I sincerely thank Science for its open access policy provided to its wide readership. This policy allows readers like me to download, read, analyze, and share this outstanding research achievement without any fees. By doing so, Science not only significantly promotes the dissemination of scientific knowledge but also opens a precious resource of knowledge to researchers, students, and those passionate about science worldwide. The concept of open access greatly advances the pace of scientific development and plays a crucial role in enhancing the public's understanding and interest in scientific work. I express my gratitude again to Science for its contribution and look forward to discovering more exciting content in the scientific world through this platform in the future.
. PDF(1016KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jia Xuan
Related articles
. Antiviral Strategy
. PLpro
. Jun12682
. Antipandemic Effects
Tools
. Email to a friend
. Post a comment